06-Jul-2022
Microtubules are core components of the cytoskeleton that are essential for cargo trafficking, cellular organisation, and force generation. They are composed of α- and β-tubulin heterodimers and are highly heterogeneous, allowing for functional specialisation. Heterogeneity is created partly due to posttranslational modifications to the C-terminal tail of α-tubulin. Specifically, the C-terminal tyrosine can be removed and religated continuously. This process is called the "tyrosination cycle" and its defects are known to lead to disease, particularly neurological problems. While religation of the tyrosine is well-understood, and happens through the tubulin tyrosine ligase (TTL), the first step of the process, detyronisation, remained poorly understood for well over four decades. To solve this puzzle, a team based at NKI, Netherlands performed genetics screens to find the enzymes involved in the process.
Four years previously, the team discovered that Vasohibins, in complex with the cofactor small vasohibin-binding protein (SVBP), was the first enzyme responsible for tubulin detyrosynation. The crystal structure of the VASH-SVBP complex revealed the enzymatic mechanism of this process, allowing new insight on the tyrosination cycle. However, cell-based experiments clearly hinted that yet another enzyme was needed for function.
The same team, with a few new members, set up to find this new, unidentified, enzyme that must play a role in the process. Using the same genetic methods, but in cells that were already missing the previously identified enzymes, Landskron et al (2022) found that the unstudied gene KIAA0895L (now MATCAP1, microtubule-associated tyrosine carboxypeptidase 1) was directly responsible for tubulin detyrosination.
The MATCAP1 activity as α tyrosine carboxypeptidase was confirmed in cells, whereby combined ablation of MATCAP1 reduced detyrosination, while overexpression of MATCAP stimulated tubulin detyrosination. Combined deletion of MATCPAP1 and vasohibins resulted in undetectable levels of detyrosination in HeLa cells, which normally have low levels of tubulin detyrosination. that the crystal structure of MATCAP1 showed that it belongs to the gluzincin metallopetidase clan, having a distinctively different enzymatic mechanism compared to vasohibins. Molecular docking studies, mutagenesis and mass spectrometry analysis revealed that MATCAP1 recognises the tubulin tail substrate in a different manner compared to the VASH1/SVBP complex
To determine how exactly MATCAP1 engages with microtubules, the team used Cryo-EM. Using the microscope at NeCEN at Leiden, an Instruct-NL facility, they collected high quality data that allowed the determination of a CryoEM structure. The structure indicated three primary binding domains; the tail binding pocket where the tyrosine is cleaved off (1), a helix-helix interface between the last helix of tubulin and a helix of MATCAP1 (2), and a loop of MATCAP1 interacting with ‘the next’ tubulin dimer (3). Two regions of interest that could not be resolved from the Cryo-EM map were also determined; the “backside” of MATCAP (4), and the N-terminus of MATCAP (5). Figure 1A shows the Cryo-EM density of MATCAP bound to a microtubule. Figure 1B illustrates the fitted molecular structure of the 5 binding regions of MATCAP, as outlined above.
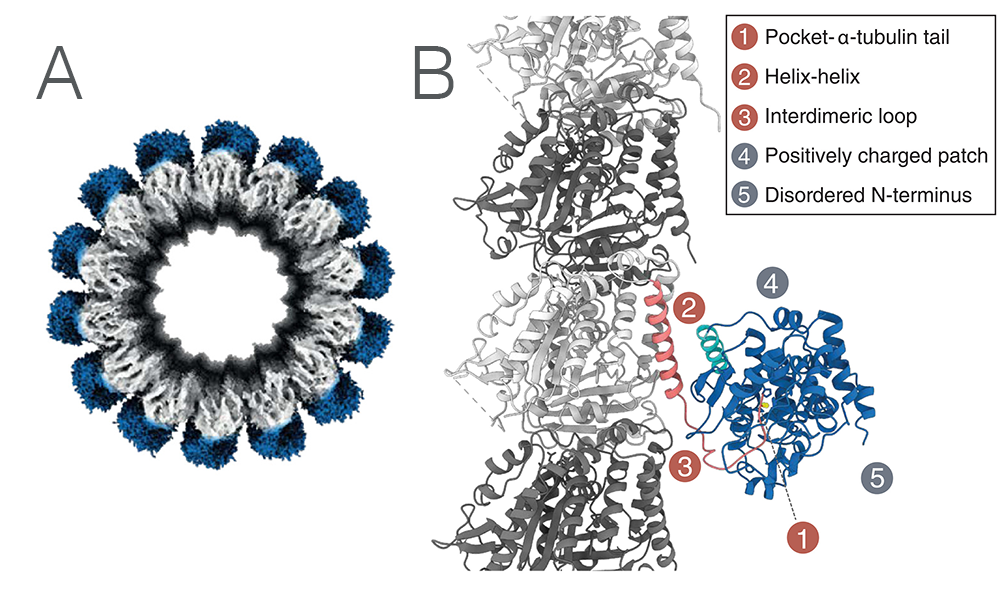
Figure 1. A: CryoEM structure of MATCAP bound to a microtubule. B: The molecular structure of MATCAP bound to α-tubulin and its five interacting regions.
To understand the contribution of each interface, the five interfaces were studied separately using mutation studies in cells. All five interfaces were indeed found to be individually important for the binding of MATCAP to the microtubules.
The study then created a detyrosination deficient mouse line by crossing MATCAP impaired and SVBP impaired mice, and surprisingly found that the offspring was viable, this in contrast with the TTL deficient mice which are lethal.
Detyrosination deficient mice had significantly reduced brain sizes, likely due to a lack of cell proliferation during embryogenesis. Mutants lacking only SVBP or MATCAP (but not both), although smaller than wild type animals, had a less pronounced effect on brain volume. An interesting behavioural feature is that the mice did not perform nesting behaviour upon having pups, indicating an important role for the detyrosinating enzymes in brain development and behaviour.
VASH-SVBP and MATCAP are totally unrelated, yet carry very similar functions – understanding their distinct structure and function could glean ongoing developments for understanding the microtubule detyrosination cycle.
Read the full text of the article here: Landskron et al., Posttranslational modification of microtubules by the MATCAP detyrosinase. Science (2022). https://doi.org/10.1126/science.abn6020
The work was also presented at the Instruct Biennial Structural Biology Conference – view the video here!