Structural biology techniques used to benefit bees and beyond
Just like other animals, bees can contract viruses from parasites such as mites. A virus of particular significance is the Deformed Wing Virus (DWV), which has been causing epidemics in Western honey bee populations. As the name suggests, DWV causes pupae to develop with deformed wings - but it can also lead to other defects, including a shortened abdomen and cuticle discolouration in adult bees. Bees with DWV often die soon after pupation, causing colonies to collapse. In 2017, Dr Nováček of the Central European Institute of Technology (CEITEC) in the Czech Republic helped to analyse data to create a detailed 3D picture of the DWV structure.
To develop a treatment for the Deformed Wing Virus, scientists first need to identify a target - this is where cryo-electron microscopy and Dr Nováček come in. By producing an atomic-scale model of the virus, cryo-EM can be used to identify the best molecular structures for a therapeutic target. The team of researchers not only ruled out some parts of the DWV structure based on their chemical properties, but also identified a potential site for treatment: the ‘P domain’. As this domain is needed for the virus to enter a host cell, it is a suitable target for an antiviral treatment.
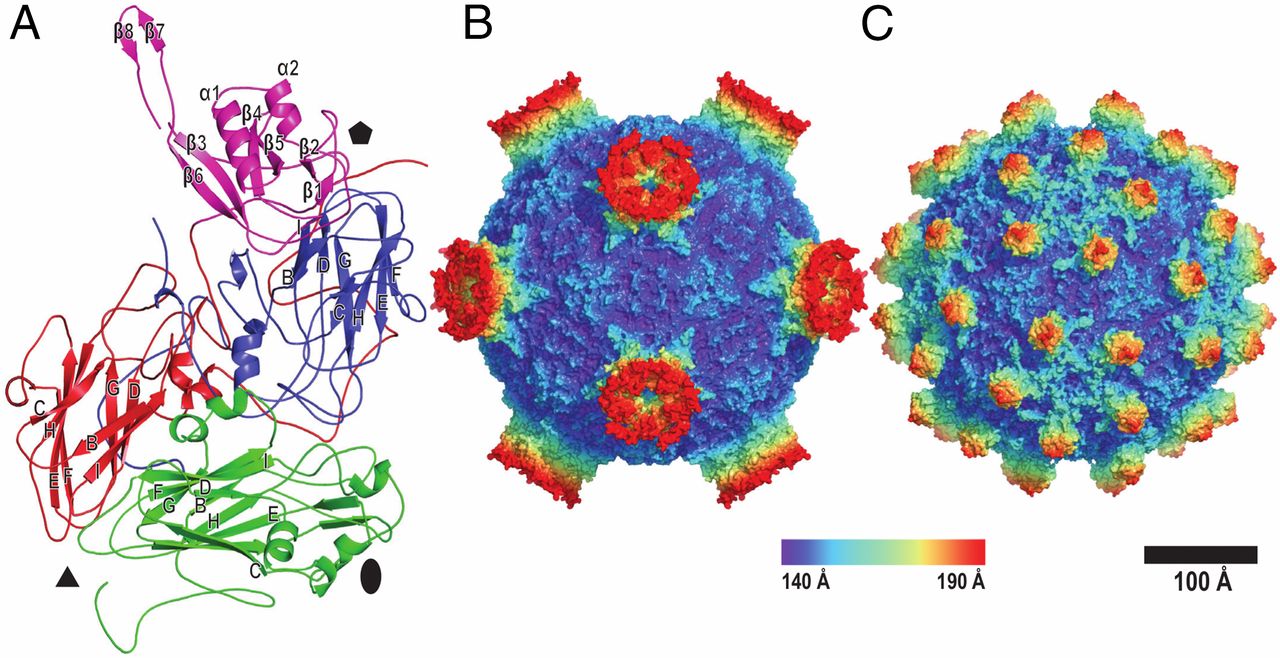
Image A shows the P domain, which is part of VP3, highlighted in magenta. Image B and C show the molecular surfaces of DWV virions determined by cryo-EM (B) and X-ray crystallography (C). (Source: Škubník K, et al. (2017) Structure of deformed wing virus, a major honey bee pathogen. Proc Natl Acad Sci USA 114:3210–3215 https://doi.org/10.1073/pnas.1615695114)
Interpreting information about the structure of biological molecules often requires a lot of time from experts. As well as cryo-EM, scientists also use nuclear magnetic resonance (NMR) spectroscopy to determine the location of atoms in a molecule and generate a model. NMR is relatively straightforward when studying small molecules, but larger molecules (like proteins) can require up to 10 different readings to be analysed by an expert.
Researchers at CEITEC, including Dr Nováček, have published an algorithm called “4D-CHAINS” to address the challenges of interpreting NMR for proteins. In their paper, the authors conclude that the algorithm “has a minimal impact on the precision and quality of the resulting structural ensembles, while allowing for a tremendous reduction in human effort and NMR spectrometer time”. By focussing on the application and development of techniques, Dr Nováček has had a hand in many scientific developments with potential benefits in a multitude of disciplines.
Read the full Deformed Wing Virus paper: Karel Škubník K., et al., 2017, Structure of deformed wing virus, a major honey bee pathogen, Proceedings of the National Academy of Sciences, 114 (12) 3210-3215
Read the full NMR algorithm paper: Evangelidis, T., et al., 2018, Automated NMR resonance assignments and structure determination using a minimal set of 4D spectra, Nature Communications, 9 (384)
Read more about NMR facilities that are accessible through Instruct-ERIC.
