HADDOCK screen of 2000 approved drugs for COVID-19
Scientists at the Instruct Centre NL have led efforts to use a software tool called HADDOCK for a large drug screen in the search for new antivirals against COVID-19. HADDOCK (High Ambiguity Driven protein-protein DOCKing) was developed by Prof Alexander Bonvin at Utrecht University as a modelling tool to predict protein-protein interactions within biomolecular complexes.
One potential drug target for the COVID-19 virus is the SARS-CoV-2 protease, which has an essential role in processing the polyproteins that are translated from the viral RNA during replication. Therefore, a SARS-CoV-2 protease inhibitor could make for an effective anticoronaviral drug. Due to the urgent need for anti-COVID-19 therapies, researchers decided to screen for drugs that are already approved for other diseases. The strategy to repurpose existing drugs for new diseases is very appealing, as these drugs have already been fully tested in clinical trials to make sure that there is no toxicity or concerning side effects.
Using a platform such as HADDOCK, allows for rapid screening of thousands of chemical compounds against the protease structure, compared to reviewing compounds one-by-one after co-crystallisation or by Cryo-EM. In the first 24 hours, for example, an average of 731 jobs were completed per hour using HADDOCK.
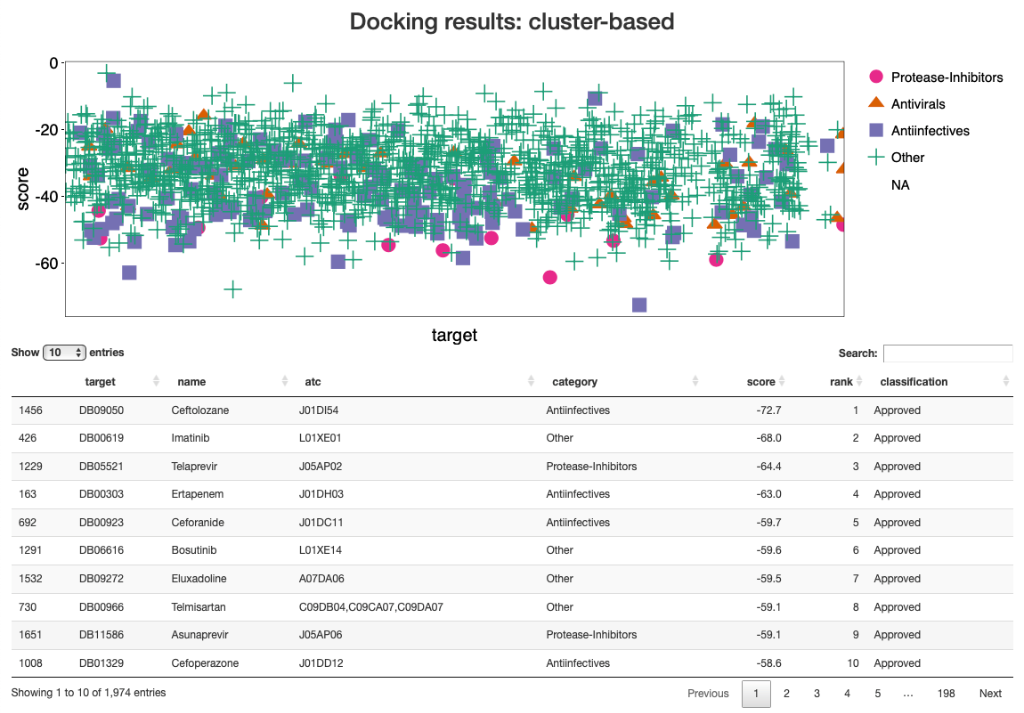
Figure 1. View of the HADDOCK results showing in the top plot the score of the various drug compounds docked against the protease and in the table below it the top10 hits. Results are accessible online at http://www.bonvinlab.org/covid.
These studies were headed up by Panagiotis Koukos and Manon Réau in collaboration with the EGI Foundation who provided advanced computing services. It is hoped that hits identified by HADDOCK modelling will lead to further research to determine the clinical efficacy of protease inhibitors as antivirals against COVID-19.
HADDOCK is a core software in the BioExcel Center of Excellence and BioExcel has also committed to support COVID-19 researchers. In recent weeks, the HADDOCK web portal has registered many users wanting to use the platform for COVID-19 projects; in the week from 10 -17 April 2020, over 70 % of HADDOCK jobs were related to COVID-19. To support collaborative projects, the HADDOCK team, together with EGI/EOSC experts, are expanding the processing capacity of the HADDOCK portals and providing customised solutions to support researchers.
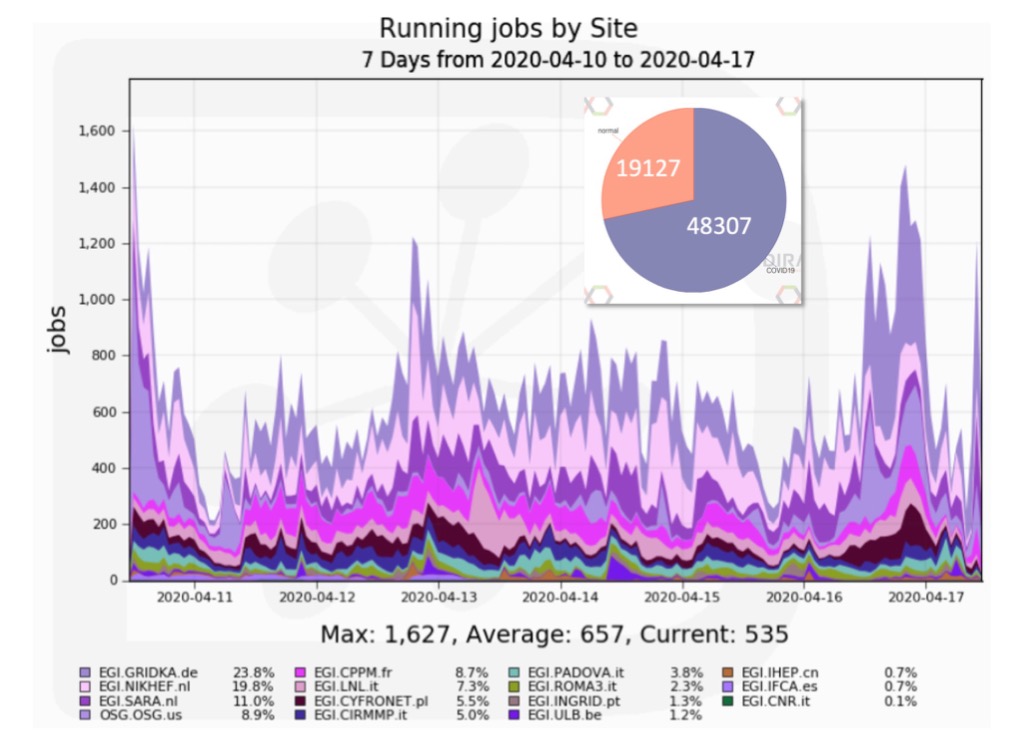
Figure 2. The number of HADDOCK jobs submitted via DIRAC4EGI to EOSC/EGI HTC resources over a one week period. The inset shows the fraction of COVID-related jobs (~72%). The running jobs are coloured based on the site on which they have been running, nicely showing how international resources, within Europe, but also the US (OSG) and China (IHEP) are being used to support the HADDOCK WeNMR portal.
For more information about HADDOCK and other protein modelling tools, consult the Instruct-ERIC Computational Services webpage.
To apply to use Instruct-ERIC Structural Biology services for COVID-19 research see our Resources for COVID-19 research.

